Tree crown detection using DeepForest
Contents
Tree crown detection using DeepForest¶
Forest Modelling
Context¶
Purpose¶
Detect tree crown using a state-of-art Deep Learning model for object detection.
Modelling approach¶
A prebuilt Deep Learning model, named DeepForest, is used to predict individual tree crowns from an airborne RGB image. DeepForest was trained on data from the National Ecological Observatory Network (NEON). DeepForest was implemented in Python 3.7 using initally Tensorflow v1.14 but later moved to Pytorch. Further details can be found in the package documentation.
Highlights¶
Fetch and load a NEON image from a Zenodo repository using
intakeanddask.Retrieve and plot the ground-truth annotations (bounding boxes) for the target image.
Load and use a pretrained DeepForest model to generate predictions from the full-image or tile-wise prediction.
Indicate the pros and cons of full-image and tile-wise prediction.
Contributions¶
Notebook¶
Modelling codebase¶
Ben Weinstein (maintainer & developer), University of Florida, @bw4sz
Henry Senyondo (support maintainer), University of Florida, @ethanwhite
Ethan White (PI and author), University of Florida, @weecology
Other contributors are listed in the GitHub repo
Modelling publications¶
Ben G Weinstein, Sergio Marconi, Mélaine Aubry-Kientz, Gregoire Vincent, Henry Senyondo, and Ethan P White. Deepforest: a python package for rgb deep learning tree crown delineation. Methods in Ecology and Evolution, 11:1743–1751, 2020. URL: https://besjournals.onlinelibrary.wiley.com/doi/abs/10.1111/2041-210X.13472, doi:https://doi.org/10.1111/2041-210X.13472.
Ben G Weinstein, Sergio Marconi, Stephanie Bohlman, Alina Zare, and Ethan White. Individual tree-crown detection in rgb imagery using semi-supervised deep learning neural networks. Remote Sensing, 2019. URL: https://www.mdpi.com/2072-4292/11/11/1309, doi:10.3390/rs11111309.
Ben G Weinstein, Sergio Marconi, Stephanie A Bohlman, Alina Zare, and Ethan P White. Cross-site learning in deep learning rgb tree crown detection. Ecological Informatics, 56:101061, 2020. URL: https://www.sciencedirect.com/science/article/pii/S157495412030011X, doi:https://doi.org/10.1016/j.ecoinf.2020.101061.
Modelling funding¶
TBD
Note
The author acknowledges DeepForest contributors. Some code snippets were extracted from DeepForest GitHub public repository.
Install and load libraries¶
!pip -q install git+https://github.com/ESM-VFC/intake_zenodo_fetcher.git ##Intake Zenodo Fetcher
!pip -q install pycurl
!pip -q install torchvision==0.10.0
!pip -q install torch==1.9.0
!pip -q install DeepForest
import glob
import os
import urllib
import numpy as np
import intake
from intake_zenodo_fetcher import download_zenodo_files_for_entry
import matplotlib.pyplot as plt
import xmltodict
import cv2
import tempfile
import torch
import warnings
warnings.filterwarnings(action='ignore')
%matplotlib inline
Set project structure¶
notebook_folder = '../modelling/forest-modelling-treecrown_deepforest'
if not os.path.exists(notebook_folder):
os.makedirs(notebook_folder)
Fetch a RGB image from Zenodo¶
Fetch a sample image from a publically accessible location.
# set catalogue location
catalog_file = os.path.join(notebook_folder, 'catalog.yaml')
with open(catalog_file, 'w') as f:
f.write('''
sources:
NEONTREE_rgb:
driver: xarray_image
description: 'NeonTreeEvaluation RGB images (collection)'
metadata:
zenodo_doi: "10.5281/zenodo.3459803"
args:
urlpath: "{{ CATALOG_DIR }}/NEONsample_RGB/2018_MLBS_3_541000_4140000_image_crop.tif"
''')
Load an intake catalog for the downloaded data.
cat_tc = intake.open_catalog(catalog_file)
for catalog_entry in list(cat_tc):
download_zenodo_files_for_entry(
cat_tc[catalog_entry],
force_download=False
)
Load sample image¶
Here we use intake to load the image through dask.
tc_rgb = cat_tc["NEONTREE_rgb"].to_dask()
tc_rgb
<xarray.DataArray (y: 1864, x: 1429, channel: 3)> dask.array<xarray-<this-array>, shape=(1864, 1429, 3), dtype=uint8, chunksize=(1864, 1429, 3), chunktype=numpy.ndarray> Coordinates: * y (y) int64 0 1 2 3 4 5 6 7 ... 1857 1858 1859 1860 1861 1862 1863 * x (x) int64 0 1 2 3 4 5 6 7 ... 1422 1423 1424 1425 1426 1427 1428 * channel (channel) int64 0 1 2
- y: 1864
- x: 1429
- channel: 3
- dask.array<chunksize=(1864, 1429, 3), meta=np.ndarray>
Array Chunk Bytes 7.62 MiB 7.62 MiB Shape (1864, 1429, 3) (1864, 1429, 3) Count 1 Tasks 1 Chunks Type uint8 numpy.ndarray - y(y)int640 1 2 3 4 ... 1860 1861 1862 1863
array([ 0, 1, 2, ..., 1861, 1862, 1863])
- x(x)int640 1 2 3 4 ... 1425 1426 1427 1428
array([ 0, 1, 2, ..., 1426, 1427, 1428])
- channel(channel)int640 1 2
array([0, 1, 2])
Load and prepare labels¶
filenames = glob.glob(os.path.join(notebook_folder, './NEONsample_RGB/*.tif'))
filesn = [os.path.basename(i) for i in filenames]
##Create ordered dictionary of .xml annotation files
def loadxml(imagename):
imagename = imagename.replace('.tif','')
fullurl = "https://raw.githubusercontent.com/weecology/NeonTreeEvaluation/master/annotations/" + imagename + ".xml"
file = urllib.request.urlopen(fullurl)
data = file.read()
file.close()
data = xmltodict.parse(data)
return data
allxml = [loadxml(i) for i in filesn]
# function to extract bounding boxes
def extractbb(i):
bb = [f['bndbox'] for f in allxml[i]['annotation']['object']]
return bb
bball = [extractbb(i) for i in range(0,len(allxml))]
print(len(bball))
1
Visualise image and labels¶
# function to plot images
def cv2_imshow(a, **kwargs):
a = a.clip(0, 255).astype('uint8')
# cv2 stores colors as BGR; convert to RGB
if a.ndim == 3:
if a.shape[2] == 4:
a = cv2.cvtColor(a, cv2.COLOR_BGRA2RGBA)
else:
a = cv2.cvtColor(a, cv2.COLOR_BGR2RGB)
return plt.imshow(a, **kwargs)
image = tc_rgb
# plot predicted bbox
image2 = image.values.copy()
target_bbox = bball[0]
print(type(target_bbox))
print(target_bbox[0:2])
<class 'list'>
[OrderedDict([('xmin', '1377'), ('ymin', '697'), ('xmax', '1429'), ('ymax', '752')]), OrderedDict([('xmin', '787'), ('ymin', '232'), ('xmax', '811'), ('ymax', '256')])]
for row in target_bbox:
cv2.rectangle(image2, (int(row["xmin"]), int(row["ymin"])), (int(row["xmax"]), int(row["ymax"])), (0, 0, 0), thickness=10, lineType=cv2.LINE_AA)
plt.figure(figsize=(15,15))
cv2_imshow(np.flip(image2,2))
plt.show()

Load DeepForest pretrained model¶
Now we’re going to load and use a pretrained model from the deepforest package.
from deepforest import main
# load deep forest model
model = main.deepforest()
model.use_release()
model.current_device = torch.device("cpu")
Reading config file: /Users/acoca/anaconda3/envs/envds-book/lib/python3.8/site-packages/deepforest/data/deepforest_config.yml
Model from DeepForest release https://github.com/weecology/DeepForest/releases/tag/1.0.0 was already downloaded. Loading model from file.
Loading pre-built model: https://github.com/weecology/DeepForest/releases/tag/1.0.0
pred_boxes = model.predict_image(image=image.values)
print(pred_boxes.head(5))
xmin ymin xmax ymax label score
0 1258.0 561.0 1399.0 698.0 Tree 0.415253
1 1119.0 527.0 1255.0 660.0 Tree 0.395937
2 7.0 248.0 140.0 395.0 Tree 0.376462
3 444.0 459.0 575.0 582.0 Tree 0.355283
4 94.0 149.0 208.0 260.0 Tree 0.347175
image3 = image.values.copy()
for index, row in pred_boxes.iterrows():
cv2.rectangle(image3, (int(row["xmin"]), int(row["ymin"])), (int(row["xmax"]), int(row["ymax"])), (0, 0, 0), thickness=10, lineType=cv2.LINE_AA)
plt.figure(figsize=(15,15))
cv2_imshow(np.flip(image3,2))
plt.show()

Comparison full image prediction and reference labels¶
Let’s compare the labels and predictions over the tested image.
fig = plt.figure(figsize=(15,15))
ax1 = plt.subplot(1, 2, 1), cv2_imshow(np.flip(image2,2))
ax2 = plt.subplot(1, 2, 2), cv2_imshow(np.flip(image3,2))
plt.show() # To show figure
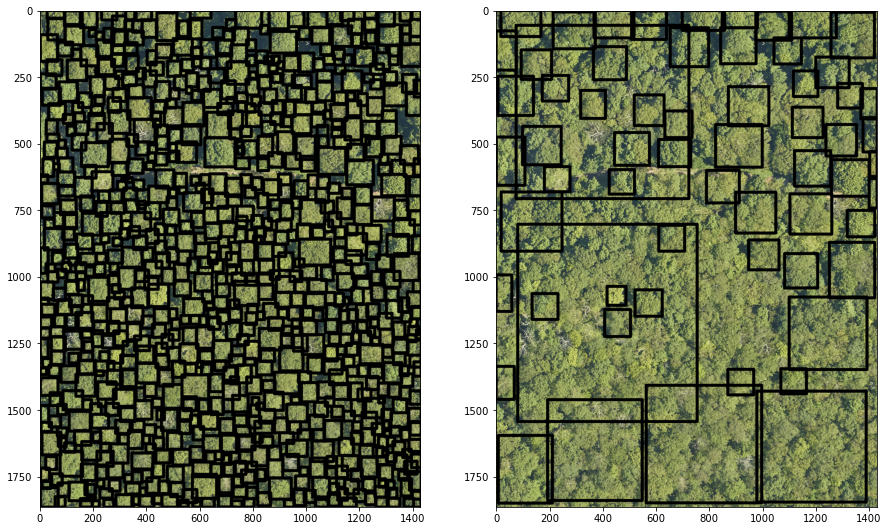
Interpretation:
It seems the pretrained model doesn’t perform well with the tested image.
The low performance might be explained due to the pretrained model used 10cm resolution images.
Tile-based prediction¶
To optimise the predictions, the DeepForest can be run tile-wise.
The following cells show how to define the optimal window i.e. tile size.
from deepforest import preprocess
#Create windows of 400px
windows = preprocess.compute_windows(image.values, patch_size=400,patch_overlap=0)
print(f'We have {len(windows)} in the image')
We have 20 in the image
#Loop through a few sample windows, crop and predict
fig, axes, = plt.subplots(nrows=2,ncols=2, figsize=(15,15))
axes = axes.flatten()
for index2 in range(4):
crop = image.values[windows[index2].indices()]
#predict in bgr channel order, color predictions in red.
boxes = model.predict_image(image=np.flip(crop[...,::-1],2), return_plot = True)
#but plot in rgb channel order
axes[index2].imshow(boxes[...,::-1])
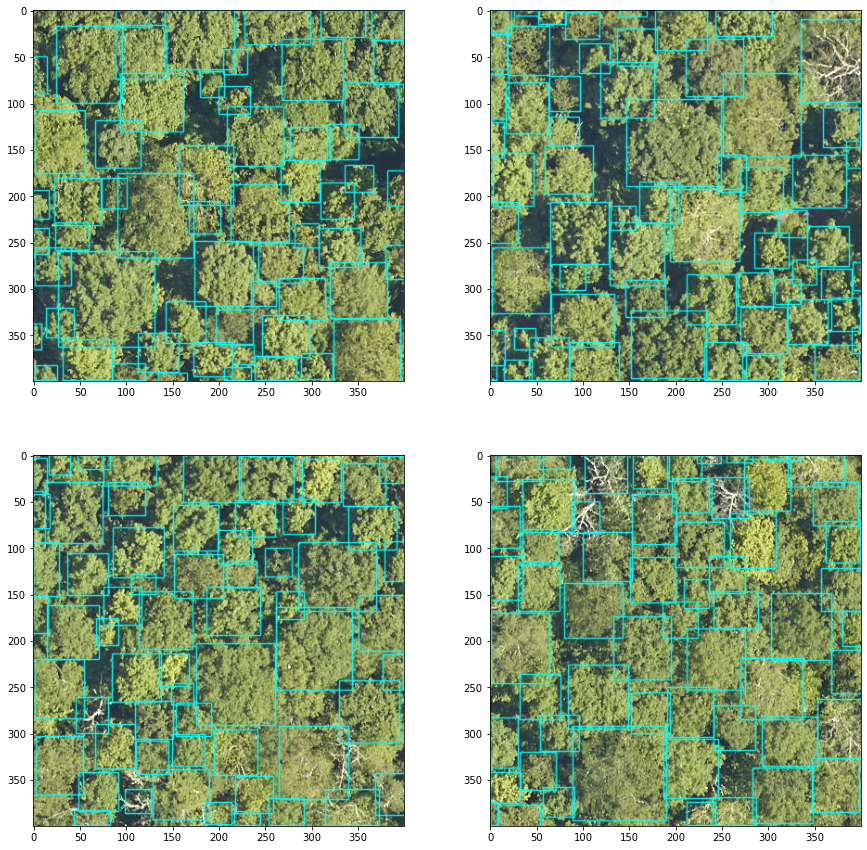
Once a suitable tile size is defined, we can run in a batch using the predict_tile function:
tile = model.predict_tile(image=image.values,return_plot=False,patch_overlap=0,iou_threshold=0.05,patch_size=400)
# plot predicted bbox
image_tile = image.values.copy()
for index, row in tile.iterrows():
cv2.rectangle(image_tile, (int(row["xmin"]), int(row["ymin"])), (int(row["xmax"]), int(row["ymax"])), (0, 0, 0), thickness=10, lineType=cv2.LINE_AA)
fig = plt.figure(figsize=(15,15))
ax1 = plt.subplot(1, 2, 1), cv2_imshow(np.flip(image2,2))
ax2 = plt.subplot(1, 2, 2), cv2_imshow(np.flip(image_tile,2))
plt.show() # To show figure
100%|██████████| 20/20 [03:21<00:00, 10.09s/it]
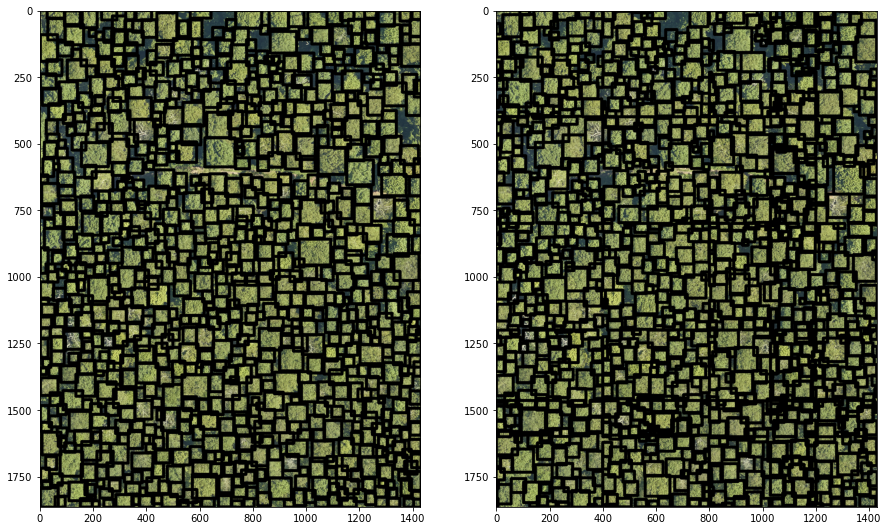
Interpretation
The tile-based prediction provides more reasonable results than predicting over the whole image.
While the prediction looks closer to the ground truth labels, there seem to be some tiles edges artefacts. This will require further investigation i.e. inspecting the
deepforesttile-wise prediction function to understand how the predictions from different tiles are combined after the model has made them.
Summary¶
This notebook has demonstrated the use of:
The
deepforestpackage to easily load and run a pretrained model for tree crown classification from very-high resolution RGB imagery.tile-wiseto considerably improve the prediction. However, user should define an optimal tile size.cv2to visualise the bounding box.